
Quick start guide
Martin Westgate & Dax Kellie
2026-02-11
Source:vignettes/quick_start_guide.Rmd
quick_start_guide.Rmdgalah is an R interface to biodiversity data hosted by
the Global Biodiversity Information Facility (GBIF) and its subsidiary node
organisations. GBIF and its partner nodes collate and store observations
of individual life forms using the ‘Darwin Core’ data standard.
Installation
To install from CRAN:
install.packages("galah")Or install the development version from GitHub:
install.packages("remotes")
remotes::install_github("AtlasOfLivingAustralia/galah")Load the package
Configuration
Begin by choosing which organisation you would like
galah to query, and providing your registration information
for that organisation.
galah_config(atlas = "GBIF",
username = "user1",
email = "email@email.com",
password = "my_password")The full list of supported queries by organisation is as follows:
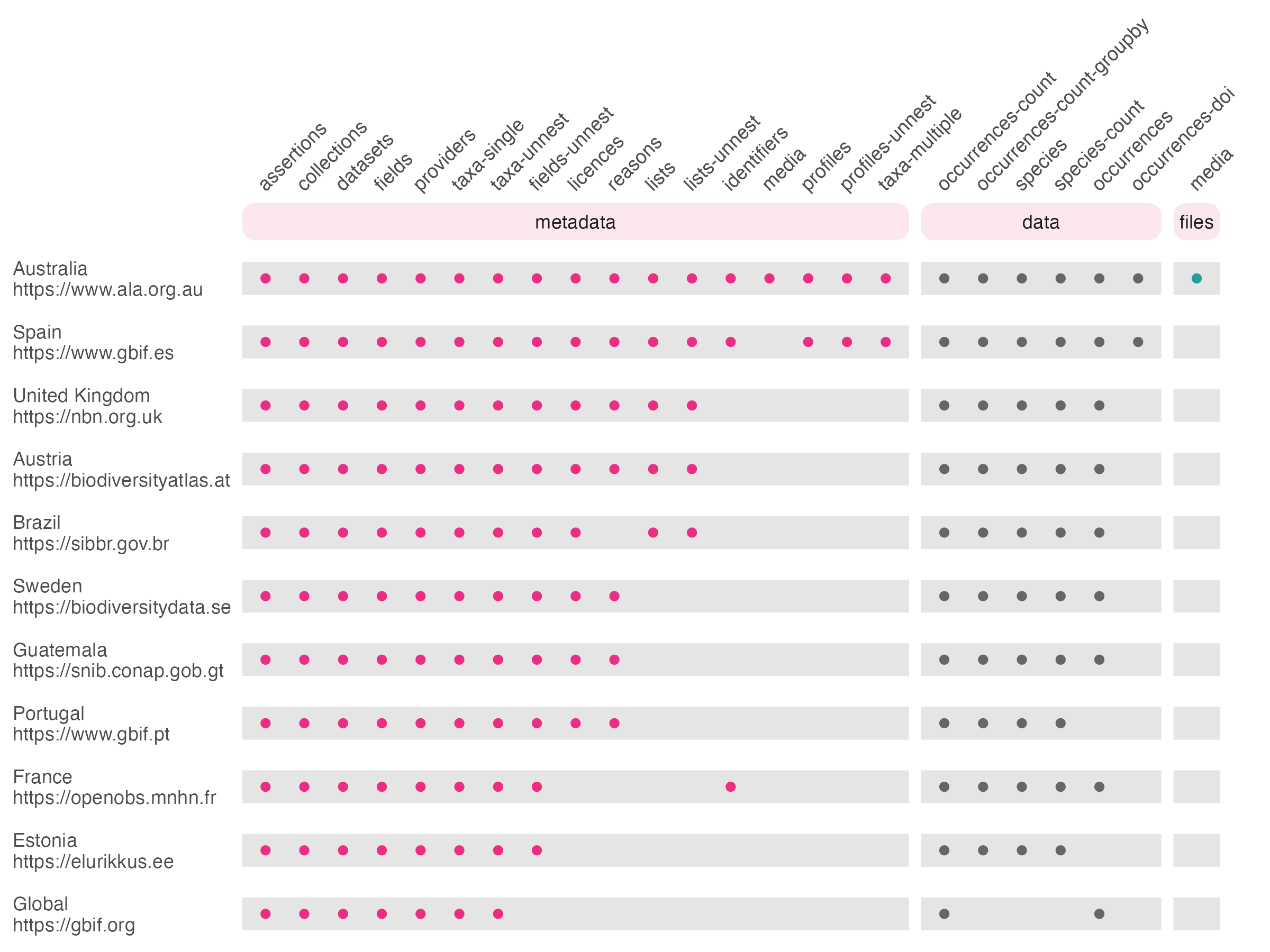
Fig 1: Organisations and APIs supported by galah
Getting data
galah is a dplyr extension package; rather
than using pipes to amend a tibble in your workspace, you
amend a query, which is then sent to your chosen organisation. These
pipes differ from traditional syntax in two ways:
- they begin with a function - usually
galah_call()- instead of atibble - they end with one of
dplyr’s evaluation functions, usuallycollect()
So an example query might be to find the number of records per year:
galah_config(atlas = "Australia")
galah_call() |> # open a pipe
filter(year >= 2020) |> # choose rows to keep
count(year) |> # count the number of rows
collect() # retrieve query from the server## # A tibble: 7 × 2
## year count
## <chr> <int>
## 1 2024 11889930
## 2 2023 11007491
## 3 2022 9430065
## 4 2025 9142677
## 5 2021 8695248
## 6 2020 7311836
## 7 2026 309836Or to find the number of categories present in a dataset, for example how many species are present:
galah_call() |>
identify("Crinia") |> # filters by taxonomic names
distinct(speciesID) |> # keep only unique values
count() |>
collect()## # A tibble: 1 × 1
## count
## <int>
## 1 17You can ‘glimpse’ a data download before you run it, to check all the data you need is included:
galah_call() |>
identify("Eolophus roseicapilla") |>
filter(year == 2010) |>
glimpse() |>
collect()## Rows: 21,984
## Columns: 8
## $ taxonConceptID <chr> "https://biodiversity.org.au/afd/taxa/9b4ad548-8bb3-486a-ab0a-905506c463ea", "https://biodiversity.org.au…
## $ eventDate <dbl> 1.272672e+12, 1.289002e+12, 1.291014e+12
## $ scientificName <chr> "Eolophus roseicapilla", "Eolophus roseicapilla", "Eolophus roseicapilla"
## $ decimalLatitude <dbl> -25.98833, -37.83032, -35.41707
## $ decimalLongitude <dbl> 152.0442, 144.9812, 138.6868
## $ basisOfRecord <chr> "HUMAN_OBSERVATION", "HUMAN_OBSERVATION", "HUMAN_OBSERVATION"
## $ dataResourceName <chr> "BirdLife Australia, Birdata", "eBird Australia", "eBird Australia"
## $ occurrenceStatus <chr> "PRESENT", "ABSENT", "ABSENT"And, once satisfied that your parameters are correct, download the records themselves:
galah_call() |>
identify("Eolophus roseicapilla") |>
filter(year == 2010) |>
select(eventDate, decimalLatitude, species) |>
collect()## # A tibble: 21,984 × 3
## eventDate decimalLatitude species
## <dttm> <dbl> <chr>
## 1 NA -36.5 Eolophus roseicapilla
## 2 NA -38.2 Eolophus roseicapilla
## 3 NA -37.0 Eolophus roseicapilla
## 4 NA -37.7 Eolophus roseicapilla
## 5 NA -35.6 Eolophus roseicapilla
## 6 NA -31.1 Eolophus roseicapilla
## 7 NA -38.2 Eolophus roseicapilla
## 8 NA -38.2 Eolophus roseicapilla
## 9 NA -38.2 Eolophus roseicapilla
## 10 NA -38.2 Eolophus roseicapilla
## # ℹ 21,974 more rowsThis works because many of the functions in dplyr are
“generic”, meaning it is possible to write extensions that apply them to
new object classes. In our case, galah_call() creates a new
object class called a data_request for which we have
written new extensions. This means that galah will not interfere with
your use of filter() and friends on your tibbles. Supported
dplyr verbs that modify queries are as follows:
arrange.data_request()count.data_request()distinct.data_request()filter.data_request()glimpse.data_request()group_by.data_request()select.data_request()slice_head.data_request()
Additional verbs are:
It is good practice to download your data in as few steps as possible, to minimize impacts on the server, and to ensure you can get a single DOI for your data. See the download data reproducibly vignette for details.
Finding information
Building queries using filter() requires that you know
two things:
- what fields (columns) are present in the dataset you are searching
- what values exist for those fields
Finding this information requires looking for metadata:
request_metadata(type = "fields") |>
collect()## # A tibble: 639 × 3
## id description type
## <chr> <chr> <chr>
## 1 abcdTypeStatus <NA> fields
## 2 acceptedNameUsage Accepted name fields
## 3 acceptedNameUsageID Accepted name fields
## 4 accessRights Access rights fields
## 5 annotationsDoi <NA> fields
## 6 annotationsUid Referenced by publication fields
## 7 assertionUserId Assertions by user fields
## 8 assertions Record issues fields
## 9 assertionsCount <NA> fields
## 10 associatedMedia Associated Media fields
## # ℹ 629 more rowsYou can browser this tibble using View() or search it
using filter(). Once you have found a field that you want
to include in your query, you can find values for that field using
unnest():
request_metadata() |>
filter(fields == "cl22") |>
unnest() |>
collect()## # A tibble: 11 × 1
## cl22
## <chr>
## 1 New South Wales
## 2 Victoria
## 3 Queensland
## 4 South Australia
## 5 Western Australia
## 6 Northern Territory
## 7 Tasmania
## 8 Australian Capital Territory
## 9 Macquarie Island
## 10 Coral Sea Islands
## 11 Ashmore and Cartier IslandsDifferent types of metadata are available; see
?request_metadata for a full list.
Wrapper functions
While dplyr syntax is very flexible, there are cases
where it is easier to simply say the sort of data you want, rather than
create a database query to implement it. For this reason, several common
use cases have their own wrapper functions.
The atlas_ family of functions act like
collect(), but enforce a particular type of data to be
returned, such as record counts:
galah_call() |>
filter(year == 2025) |>
atlas_counts() # note no need for a `count()` function## # A tibble: 1 × 1
## count
## <int>
## 1 9142677Or occurrences:
galah_call() |>
identify("Eolophus roseicapilla") |>
filter(year == 2000,
cl22 == "Australian Capital Territory") |>
atlas_occurrences() |>
print(n = 6)## # A tibble: 2,032 × 9
## recordID scientificName taxonConceptID decimalLatitude decimalLongitude eventDate basisOfRecord occurrenceStatus
## <chr> <chr> <chr> <dbl> <dbl> <dttm> <chr> <chr>
## 1 0026d29f-b6ab-4… Eolophus rose… https://biodi… -35.4 149. 2000-08-07 00:00:00 HUMAN_OBSERV… PRESENT
## 2 0062d446-007b-4… Eolophus rose… https://biodi… -35.3 149. 2000-03-10 00:00:00 HUMAN_OBSERV… PRESENT
## 3 00a62ee0-1e08-4… Eolophus rose… https://biodi… -35.2 149. 2000-01-29 00:00:00 HUMAN_OBSERV… PRESENT
## 4 00ab2f4d-326f-4… Eolophus rose… https://biodi… -35.4 149. 2000-09-25 00:00:00 HUMAN_OBSERV… PRESENT
## 5 00ae4631-ea59-4… Eolophus rose… https://biodi… -35.3 149. 2000-02-12 00:00:00 HUMAN_OBSERV… PRESENT
## 6 00b6c8ec-e7b9-4… Eolophus rose… https://biodi… -35.2 149. 2000-02-05 00:00:00 HUMAN_OBSERV… PRESENT
## # ℹ 2,026 more rows
## # ℹ 1 more variable: dataResourceName <chr>atlas_species() replaces the need for
distinct() call, while atlas_media() is a
shortcut to a complex workflow that incorporates both data and metadata
calls. Finally, metadata calls can be made more efficiently using the
show_all() and show_values() functions. These
take the same arguments as the type argument in
request_metadata(), but use non-standard evaluation, so
they don’t require quotes. They are also evaluated immediately rather
than lazily:
show_all(fields)## # A tibble: 639 × 3
## id description type
## <chr> <chr> <chr>
## 1 abcdTypeStatus <NA> fields
## 2 acceptedNameUsage Accepted name fields
## 3 acceptedNameUsageID Accepted name fields
## 4 accessRights Access rights fields
## 5 annotationsDoi <NA> fields
## 6 annotationsUid Referenced by publication fields
## 7 assertionUserId Assertions by user fields
## 8 assertions Record issues fields
## 9 assertionsCount <NA> fields
## 10 associatedMedia Associated Media fields
## # ℹ 629 more rowsYou can check the look up information vignette for further details.