
Download data reproducibly
Dax Kellie & Martin Westgate
2025-02-07
Source:vignettes/download-data-reproducibly.Rmd
download-data-reproducibly.RmdReproducible data downloads are important for a project’s long-term use and longevity. Below we briefly discuss why galah queries aren’t reproducible by default, how to make a data download reproducible, and how to download the same data again in future.
The trade-off of new data
By default, galah downloads data directly from each Living Atlas API. This default means that galah automatically downloads the most up-to-date data available (yay!).
Living Atlases, however, are constantly ingesting more data. These new data may be from within the last week, the last year, or the last hundred years, depending on the source. As a result, even if we run the same query, the data we download today may return a different result from the data we downloaded yesterday! Frequent data ingestion means that although galah is useful because it downloads the latest data, galah won’t necessarily return the same data every time we run the same query. How can we preserve the result returned by a query if the query always changes?
We recommend you take two steps to ensure your data are stable:
- download data to a specified location so you can find and re-use it later
- request a DOI with your download so you can request it again if needed (and cite it correctly!)
These steps are independent, so you can choose whether to enact none, one, or both of them in the same workflow. We will show you how to do both.
Generate a data DOI
To make data downloads reproducible, galah allows us to mint a unique DOI (Digital Object Identifier) for a specific query and its subsequent result.
A DOI works very similarly to a url, in that it holds content that is accessed using a specific link. However, a url can “break” if the url is renamed or the content once found under a certain url is moved to a new url. In contrast, a DOI is persistent and will always direct a user to the same content, even if the url changes. This persistence is why DOIs are valuable for long-term reproducibility of content.
In galah, you can also add a DOI to your download query. You just
need to add mint_doi = TRUE to
atlas_occurrences(). A unique DOI will be assigned to the
resulting object once the query is run.
Please note that as of version 2.2.0, DOI generation and reload is currently supported for queries to the Atlas of Living Australia and GBIF. Other atlases don’t support DOIs at all (yet).
library(galah)
galah_config(email = "your-email-here",
directory = "ALA_downloads")
# download bandicoot occurrence records from 2003
occs <- galah_call() |>
identify("perameles") |>
filter(year == 2003) |>
atlas_occurrences(mint_doi = TRUE, # add DOI
file = "bandicoots_2003_data.zip" # specify filename
)
occs |> print(n = 5)## # A tibble: 201 × 8
## recordID scientificName taxonConceptID decimalLatitude decimalLongitude
## <chr> <chr> <chr> <dbl> <dbl>
## 1 00300a96-e… Perameles nas… https://biodi… -33.7 151.
## 2 0134472a-2… Perameles nas… https://biodi… -34.7 151.
## 3 01e7b99e-9… Perameles gun… https://biodi… -42.9 147.
## 4 029c49f0-b… Perameles nas… https://biodi… -35.1 151.
## 5 0345dbe1-9… Perameles nas… https://biodi… -35.5 150.
## # ℹ 196 more rows
## # ℹ 3 more variables: eventDate <dttm>, occurrenceStatus <chr>,
## # dataResourceName <chr>galah preserves lots of information in an object’s attributes, many
which are used to construct the API query sent to the specified Living
Atlas. We can view the new DOI assigned to occs by checking
its attributes.
attributes(occs)$doi## [1] "https://doi.org/10.26197/ala.7605ea98-e90f-46d4-998c-1b6673c48d34"We can also view information on how to cite this dataset:
atlas_citation(occs)## The citation for this dataset is:
##
## Atlas of Living Australia (7 February 2025) Occurrence download
## https://doi.org/10.26197/ala.7605ea98-e90f-46d4-998c-1b6673c48d34
##
## Please consider citing R & galah, in addition to your dataset:
##
## R Core Team (2024). _R: A Language and Environment for Statistical
## Computing_. R Foundation for Statistical Computing, Vienna, Austria.
## <https://www.R-project.org/>.
##
## Westgate M, Kellie D, Stevenson M & Newman P (2025): _galah: Biodiversity
## Data from the GBIF Node Network_. R package version 2.1.1. doi:
## 10.32614/CRAN.package.galahIf you need to reload this data locally, you can do that by simply
calling read_zip():
## # A tibble: 201 × 8
## recordID scientificName taxonConceptID decimalLatitude decimalLongitude
## <chr> <chr> <chr> <dbl> <dbl>
## 1 00300a96-e… Perameles nas… https://biodi… -33.7 151.
## 2 0134472a-2… Perameles nas… https://biodi… -34.7 151.
## 3 01e7b99e-9… Perameles gun… https://biodi… -42.9 147.
## 4 029c49f0-b… Perameles nas… https://biodi… -35.1 151.
## 5 0345dbe1-9… Perameles nas… https://biodi… -35.5 150.
## # ℹ 196 more rows
## # ℹ 3 more variables: eventDate <dttm>, occurrenceStatus <chr>,
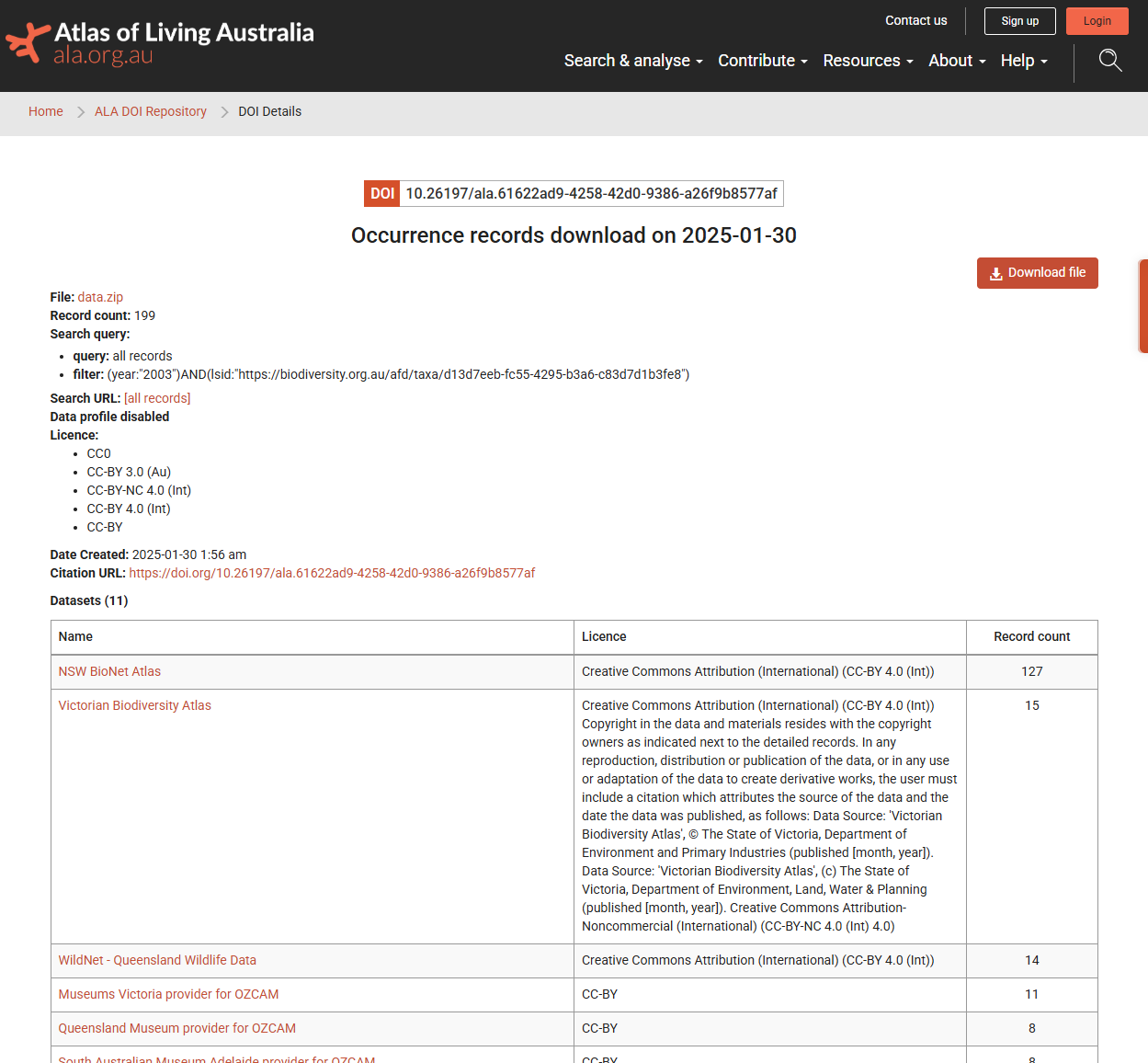
## # dataResourceName <chr>More importantly, though, we now have a persistent link to this data download. If we paste this DOI into your browser, we can see additional information about our query, including a breakdown of data providers and licensing.
plot of chunk atlas-support
Download data using a DOI
To use a DOI to return the results of a query again, we’ll use
galah_filter(). We can specify that we would like to filter
our results to only the records returned by our DOI.
occs_again <- galah_call() |>
filter(doi == attributes(occs)$doi) |> # filter by doi
atlas_occurrences()
occs_again |> print(n = 5)## # A tibble: 201 × 8
## recordID scientificName taxonConceptID decimalLatitude decimalLongitude
## <chr> <chr> <chr> <dbl> <dbl>
## 1 00300a96-e… Perameles nas… https://biodi… -33.7 151.
## 2 0134472a-2… Perameles nas… https://biodi… -34.7 151.
## 3 01e7b99e-9… Perameles gun… https://biodi… -42.9 147.
## 4 029c49f0-b… Perameles nas… https://biodi… -35.1 151.
## 5 0345dbe1-9… Perameles nas… https://biodi… -35.5 150.
## # ℹ 196 more rows
## # ℹ 3 more variables: eventDate <dttm>, occurrenceStatus <chr>,
## # dataResourceName <chr>Our query reproduces the the same records as our original query!
If you would like other examples, we use this DOI method to reproducibly download data throughout the Cleaning Biodiversity Data in R book.