Choosing an atlas
Martin Westgate & Dax Kellie
2024-11-05
Source:vignettes/choosing_an_atlas.Rmd
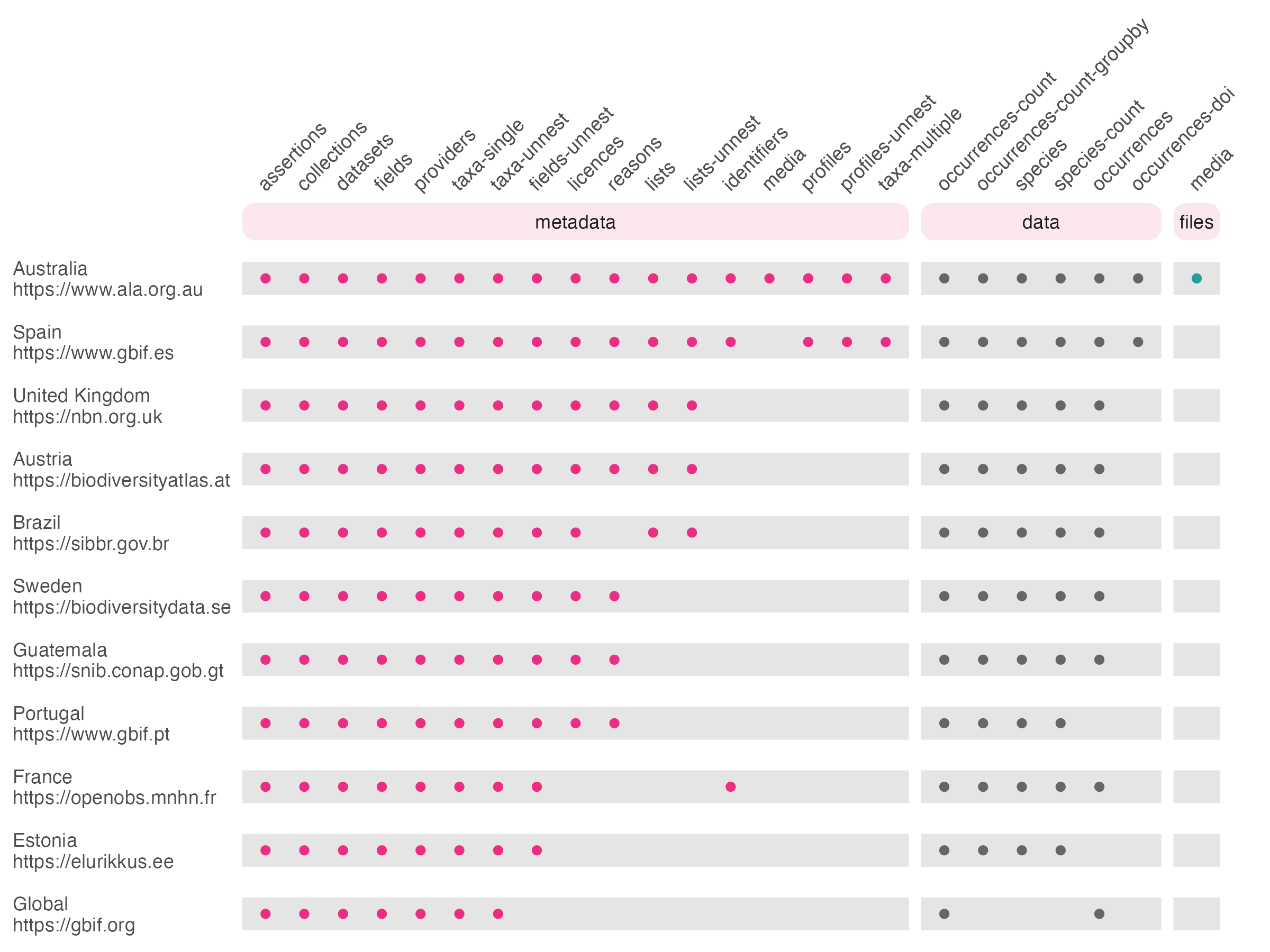
choosing_an_atlas.RmdThe GBIF network consists of a series of a series of ‘node’
organisations who collate biodiversity data from their own countries,
with GBIF acting as an umbrella organisation to store data from all
nodes. Several nodes have their own APIs, often built from the ‘living
atlas’ codebase developed by the ALA. galah enables you to
download data either from GBIF (see also rgbif) or from one
of 9 GBIF nodes:
Choosing which node to query is not entirely straightforward. Broadly speaking, GBIF is always an easy answer, because it has information from many countries. If you only want data from a single country, however, the nodes may offer some advantages. Namely, GBIF nodes may support locally-specific:
- fields that can be used for more efficient filtering
- taxonomies that reflect changes not yet adopted by the GBIF taxonomic backbone
- services that are not offered by GBIF such as error checking or large downloads
Ultimately, galah aims to provide access to as
many GBIF nodes as possible; it’s up to you which node organisation’s
data are suitable for your needs! We currently support the following
functions and atlases: