Spatial filtering#
Adapted from the Spatial Filtering galah-R article.
Callum Waite, Shandiya Balasubramaniam, Amanda Buyan
Biodiversity queries to the ALA usually require some spatial filtering. Let’s see how spatial data
are stored in the ALA and a few different methods to spatially filter data with {galah}, {shapely}
and {geopandas}.
Most records in the ALA contain location data in the form of two key fields: decimalLatitude and
decimalLongitude. As expected, these fields are the decimal coordinates of each occurrence, with south
and west values denoted by negatives. While there may be some uncertainty in these values (see field
coordinateUncertaintyInMeters), they are generally very accurate.
While these are very important and useful fields, very rarely will we encounter situations that require
queries directly calling decimalLatitude and decimalLongitude. Instead, the ALA and galah have a number
of features that make spatial queries simpler.
Contextual and spatial layers#
Often we want to filter results down to some commonly defined spatial regions, such as states, LGAs or IBRA/IMCRA regions. The ALA contains a large range (>100) of contextual and spatial layers, in-built as searchable and queriable fields. They are denoted by names beginning with “cl”, followed by an identifying number that may be up to 6 digits long. These fields are each based on shapefiles, and contain the names of the regions in these layers that each record lies in.
We strongly recommend using galah.search_all(fields="<your-search-term-here>") to check whether a contextual
layer already exists in the ALA that matches what you require before proceeding with other methods of spatial
filtering. These fields are all able to be used in the filters argument of atlas_counts, atlas_occurrences,
atlas_species or atlas_media, so they are generally easier to use.
Suppose we are interested in querying records of the Red-Necked Avocet (Recurvirostra novaehollandiae) in a particular protected wetlands, the Coorong wetlands in South Australia. We can search the ALA fields for wetlands.
>>> import galah
>>> galah.galah_config(email="your-email-here")
>>> galah.search_all(fields="wetlands")
id description type link
0 cl10829 Murray-Darling Basin Wetlands wetlands_mdb_name_only layers
1 cl10831 Murray-Darling Basin Wetland Groups wetlands_mdb_group_only layers
2 cl901 Directory of Important Wetlands Directory of Important Wetlands layers
3 cl11192 Ramsar Wetlands of Australia 2024 Ramsar_Wetlands_of_AustraliaWGS84 layers
4 cl3001 Wetlands GIS of the Murray-Darling Basin Series 2.0 Wetlands GIS of the Murray-Darling Basin Series 2.0 layers
5 cl13001 Wetlands GIS of the Murray-Darling Basin Series 2.0 Wetlands GIS of the Murray-Darling Basin Series 2.0 layers
Our search identifies that layer cl901 seems to match what we are looking for. We can then either view all
possible values in the field with galah.show_values(), or use galah.search_values() again for our particular field.
>>> galah.search_values(field="cl901",value="coorong")
field category
0 cl901 The Coorong, Lake Alexandrina & Lake Albert
We can filter all occurrences for exact matches with this value, “The Coorong, Lake Alexandrina & Lake Albert”. Our galah query can be built as follows:
>>> avocets = galah.atlas_occurrences(
... taxa = "Recurvirostra novaehollandiae",
... filters = "cl901=The Coorong, Lake Alexandrina & Lake Albert"
... )
>>> avocets.head(5)
decimalLatitude decimalLongitude eventDate scientificName taxonConceptID recordID dataResourceName occurrenceStatus
0 -36.525833 139.823608 2012-07-29T00:00:00Z Recurvirostra novaehollandiae https://biodiversity.org.au/afd/taxa/c69e7308-527a-429d-a80d-143bd20b5100 838c1162-6c3e-4b5f-84d3-3639556f1acf BirdLife Australia, Birdata PRESENT
1 -36.521950 139.827200 2023-10-17T00:00:00Z Recurvirostra novaehollandiae https://biodiversity.org.au/afd/taxa/c69e7308-527a-429d-a80d-143bd20b5100 c91d351f-06d7-4a0a-95a5-6d77e0921923 SA Fauna PRESENT
2 -36.521950 139.827200 2010-10-21T00:00:00Z Recurvirostra novaehollandiae https://biodiversity.org.au/afd/taxa/c69e7308-527a-429d-a80d-143bd20b5100 8fdebcda-de5e-4c2c-94dc-a9fb25cd6817 Australian Waterbird Surveys (1983-2018) PRESENT
3 -36.521950 139.827200 NaN Recurvirostra novaehollandiae https://biodiversity.org.au/afd/taxa/c69e7308-527a-429d-a80d-143bd20b5100 e02745af-44d4-4ad8-9816-7aaaaa318a82 Australian Waterbird Surveys (1983-2018) PRESENT
4 -36.521950 139.827200 NaN Recurvirostra novaehollandiae https://biodiversity.org.au/afd/taxa/c69e7308-527a-429d-a80d-143bd20b5100 3e4d190c-7032-48ba-885e-ebf513fe5af0 Australian Waterbird Surveys (1983-2018) PRESENT
Shapes and Regions#
While server-side spatial information is useful, there are likely to be cases where the shapefile or region
you wish to query will not be pre-loaded as a contextual layer in the ALA. In this case, shapefiles can be
introduced to the filtering process using the {shapely} package and the polygon or bbox arguments in
atlas_counts, atlas_occurrences, atlas_species or atlas_media. Shapefiles can be provided as a
shapely object, the string denoting the polygon itself, or in the base of bbox, a dictionary
specifying xmin, xmax, ymin, and ymax. For instance, we might interested in species occurrences in
King George Square, Brisbane. We can take the MULTIPOLYGON object for the square (as sourced from the Brisbane City
Council) and transform it into a {shapely} object.
>>> import shapely.wkt
>>> king_george_square = shapely.wkt.loads("MULTIPOLYGON(((153.0243 -27.46886, 153.0242 -27.46896, 153.0236 -27.46837, 153.0239 -27.46814, 153.0239 -27.46813, 153.0242 -27.46789, 153.0244 -27.46805, 153.0245 -27.46821, 153.0246 -27.46828, 153.0247 -27.46835, 153.0248 -27.46848, 153.0246 -27.4686, 153.0246 -27.46862, 153.0245 -27.46871, 153.0243 -27.46886)))")
We can provide this MULTIPOLYGON as the argument polygon in atlas_occurrences to assess which species
have been recorded in King George Square.
>>> species_king_george_square = galah.atlas_occurrences(
... polygon=king_george_square,
... fields=["decimalLatitude", "decimalLongitude", "eventDate", "scientificName", "vernacularName"]
... )
>>> species_king_george_square.head(10)
decimalLatitude decimalLongitude eventDate scientificName vernacularName
0 -27.468625 153.024333 2006-03-30T09:32:00Z Threskiornis moluccus Australian White Ibis
1 -27.468611 153.024444 2023-12-02T09:12:02Z Cosmophasis baehrae NaN
2 -27.468546 153.023911 2024-02-03T20:45:00Z Mediastinia NaN
3 -27.468529 153.023898 2025-03-19T07:01:00Z Manorina (Myzantha) melanocephala Noisy Miner
4 -27.468510 153.023821 2023-04-08T17:49:00Z Entomyzon cyanotis Blue-faced Honeyeater
5 -27.468418 153.024140 2022-08-30T11:29:00Z Threskiornis moluccus Australian White Ibis
6 -27.468364 153.024121 2024-08-28T10:31:54Z Vitellus NaN
7 -27.468326 153.024077 2022-07-24T14:43:00Z Threskiornis moluccus Australian White Ibis
8 -27.468308 153.024097 2024-10-13T14:34:00Z Threskiornis moluccus Australian White Ibis
9 -27.468295 153.023967 2024-10-13T14:35:00Z Gymnorhina tibicen Australian Magpie
10 -27.468294 153.023907 2024-11-11T13:56:00Z Threskiornis moluccus Australian White Ibis
11 -27.468262 153.023941 2025-03-19T06:55:00Z Intellagama lesueurii Eastern Water Dragon
12 -27.468167 153.024047 2024-07-23T10:00:53Z Threskiornis moluccus Australian White Ibis
13 -27.468155 153.024011 2024-04-26T12:07:00Z Threskiornis moluccus Australian White Ibis
This argument can be provided as its string; however, we show you the above to give you an idea of what
galah-python does when you provide it a string.
There is a third argument of atlas_counts, atlas_occurrences, atlas_species or atlas_media
called simplify_polygon, which defaults to False. By setting the simplify_polygon argument
to True, the provided POLYGON or MULTIPOLYGON will be converted into the smallest bounding box
(rectangle) that contains the POLYGON. In this case, records will be included that may not exactly lie
inside the provided shape.
>>> species_king_george_square = galah.atlas_occurrences(
... polygon=king_george_sq,
... simplify_polygon=True,
... fields=["decimalLatitude", "decimalLongitude", "eventDate", "scientificName", "vernacularName"]
... )
>>> species_king_george_square.head(10)
decimalLatitude decimalLongitude eventDate scientificName vernacularName
0 -27.468625 153.024333 2006-03-30T09:32:00Z Threskiornis moluccus Australian White Ibis
1 -27.468611 153.024444 2023-12-02T09:12:02Z Cosmophasis baehrae NaN
2 -27.468546 153.023911 2024-02-03T20:45:00Z Mediastinia NaN
3 -27.468529 153.023898 2025-03-19T07:01:00Z Manorina (Myzantha) melanocephala Noisy Miner
4 -27.468510 153.023821 2023-04-08T17:49:00Z Entomyzon cyanotis Blue-faced Honeyeater
5 -27.468418 153.024140 2022-08-30T11:29:00Z Threskiornis moluccus Australian White Ibis
6 -27.468364 153.024121 2024-08-28T10:31:54Z Vitellus NaN
7 -27.468326 153.024077 2022-07-24T14:43:00Z Threskiornis moluccus Australian White Ibis
8 -27.468308 153.024097 2024-10-13T14:34:00Z Threskiornis moluccus Australian White Ibis
9 -27.468295 153.023967 2024-10-13T14:35:00Z Gymnorhina tibicen Australian Magpie
Large shapefiles#
The simplify_polygon argument with option polygon is provided because objects with a large amount of
vertices will take a long time to filter on the ALA’s end. In the event you have a large shapefile, using
simplify_polygon=True will at least enable an initial reduction of the time it takes for the data to be
downloaded, before finer filtering to the actual shapefile will obtain the desired set of occurrences.
Alternatively, one can also perform the “bbox” reduction before passing the shape to atlas_counts,
atlas_occurrences, atlas_species or atlas_media by using {geopandas} and the unary_union
function of the {shapely} package.
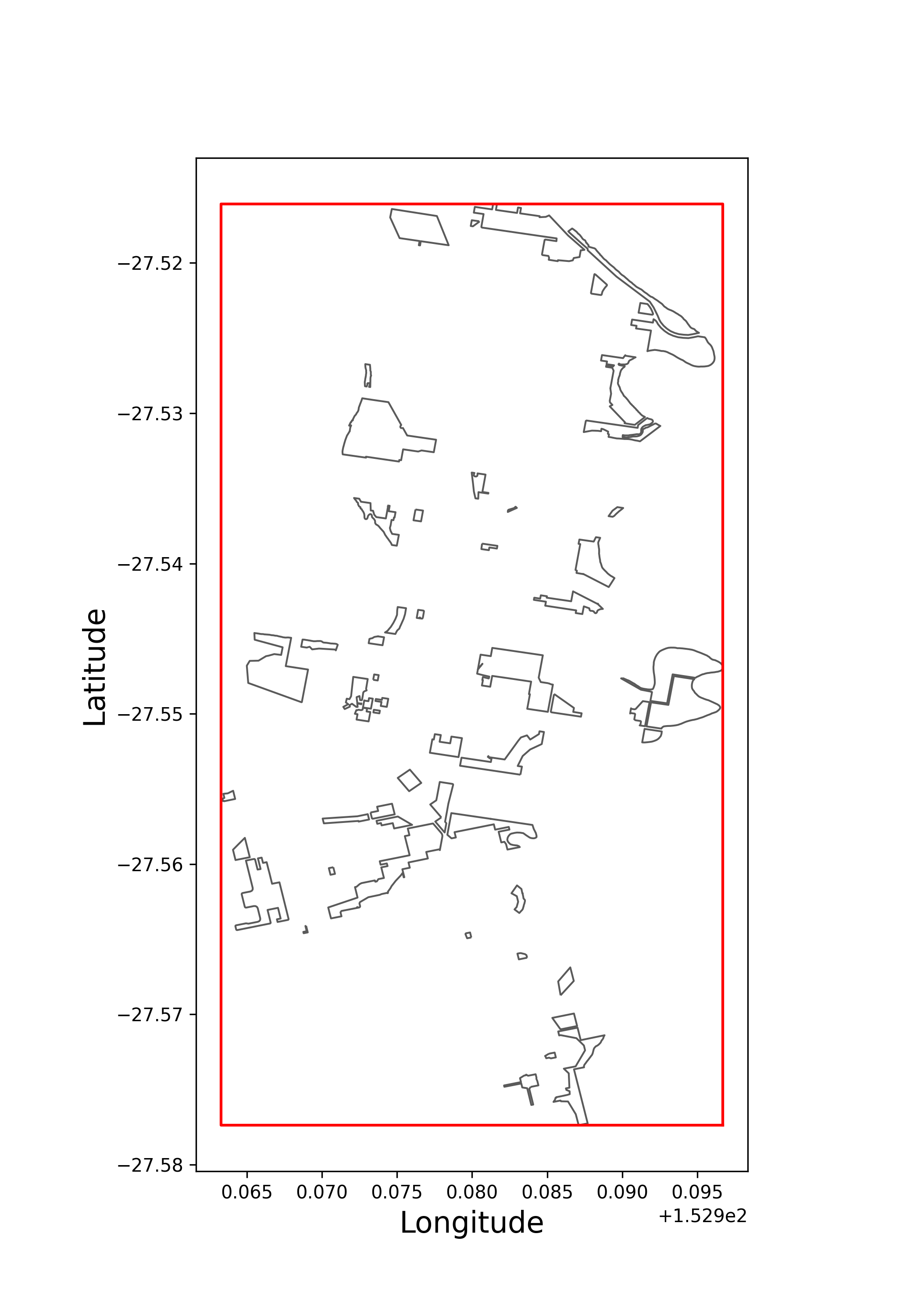
A common situation for this to occur is when a shapefile with multiple shapes is provided, where we are interested in grouping our results by each shape. Here is a mock workflow using a subset of a shapefile of all 2,184 Brisbane parks.
Let’s say we are interested in knowing which parks in the Brisbane postcode 4075 have the most occurrences of the Scaly-Breasted Lorikeet, Trichoglossus chlorolepidotus, since 2020. We can download the entire shapefile from the above link, and perform our filtering and summarising as follows:
>>> # first, get all parts within the Brisbane postcode 4075
>>> import geopandas as gpd
>>> parks = gpd.read_file("Park___Locations.shp")
>>> parks.columns
>>> brisbane_parks = parks[parks["POST_CODE"] == '4075']
>>> brisbane_parks.head(10)
OBJECTID ... geometry
0 64 ... POLYGON ((152.99236 -27.53098, 152.99118 -27.5...
1 65 ... POLYGON ((152.99137 -27.53090, 152.99130 -27.5...
2 102 ... POLYGON ((152.98534 -27.57025, 152.98534 -27.5...
3 115 ... POLYGON ((152.98702 -27.57090, 152.98723 -27.5...
4 116 ... POLYGON ((152.98525 -27.57294, 152.98513 -27.5...
5 147 ... POLYGON ((152.98423 -27.57400, 152.98429 -27.5...
6 166 ... POLYGON ((152.96348 -27.55583, 152.96348 -27.5...
7 188 ... POLYGON ((152.98312 -27.56635, 152.98302 -27.5...
8 401 ... POLYGON ((152.98792 -27.52198, 152.98800 -27.5...
9 402 ... POLYGON ((152.98165 -27.51611, 152.98159 -27.5...
[10 rows x 13 columns]
>>> # second, get a shape of all the Brisbane parks and draw a bounding box around it
>>> import shapely
>>> from shapely.ops import unary_union
>>> brisbane_parks_all = gpd.GeoSeries(unary_union(brisbane_parks["geometry"]))
>>> brisbane_parks_bbox = brisbane_parks_all.bounds
minx miny maxx maxy
0 152.963302 -27.577386 152.996692 -27.516073
To visualise
>>> plot_brisbane_parks_bbox = shapely.box(xmin=brisbane_parks_bbox["minx"][0],
... xmax=brisbane_parks_bbox["maxx"][0],
... ymin=brisbane_parks_bbox["miny"][0],
... ymax=brisbane_parks_bbox["maxy"][0]
... )
>>> brisbane_parks_all.plot(edgecolor = "#5A5A5A", linewidth = 1, facecolor = "white", figsize = (7,10))
>>> plt.plot(*plot_brisbane_parks_bbox.exterior.xy,color="red")
>>> plt.ylabel("Latitude",size=16,x=.45,y=0.5)
>>> plt.xlabel("Longitude",size=16)
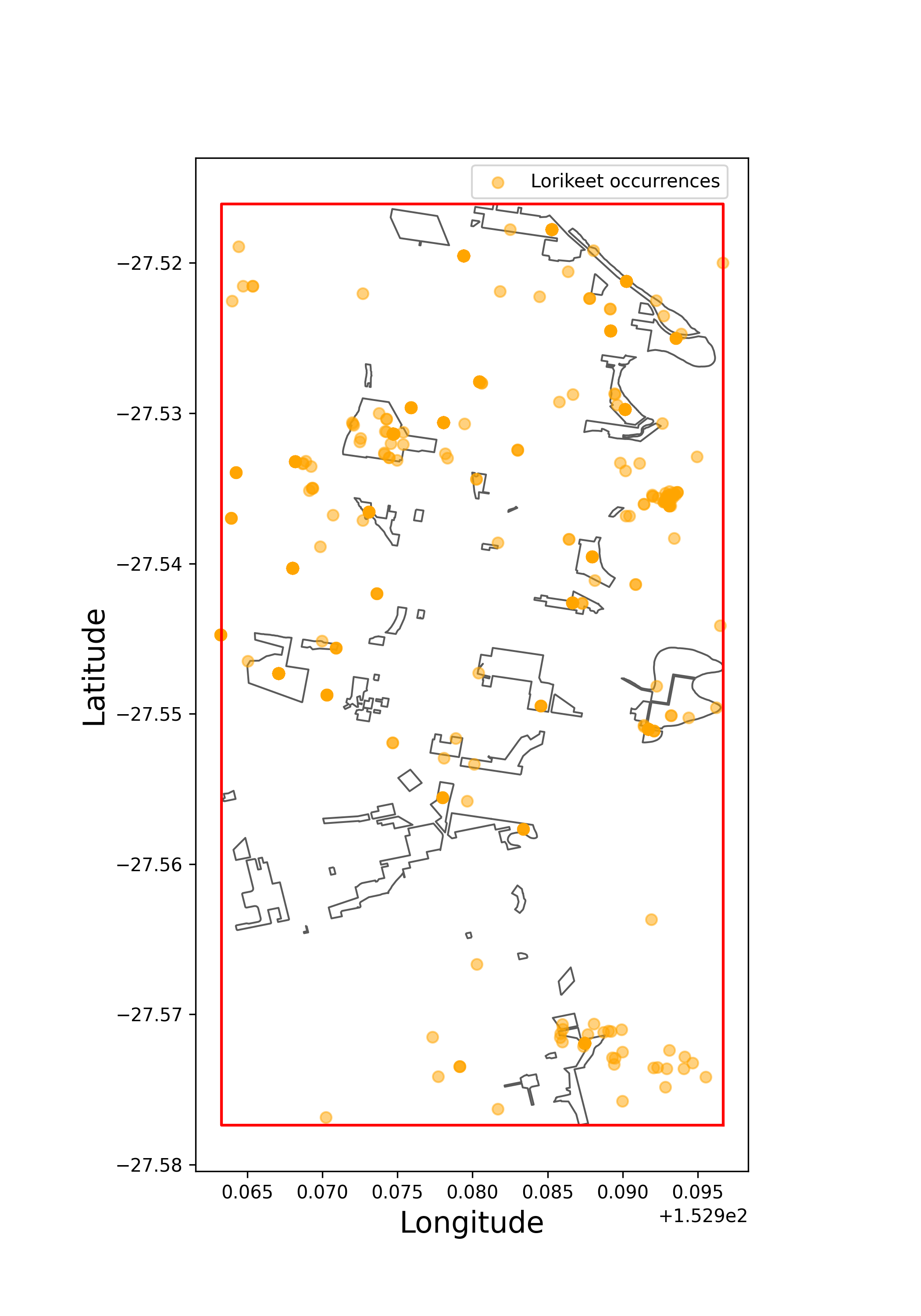
>>> # third, find all occurrences of Trichoglossus chlorolepidotus in the bounding box in 2022
>>> import galah
>>> galah.galah_config(email="your-email-here")
>>> lorikeet_brisbane = galah.atlas_occurrences(
... taxa="Trichoglossus chlorolepidotus",
... filters="year=2022",
... bbox=brisbane_parks_bbox
... )
>>> lorikeet_brisbane
decimalLatitude decimalLongitude ... dataResourceName occurrenceStatus
0 -27.576858 152.970235 ... eBird Australia PRESENT
1 -27.576303 152.981694 ... eBird Australia PRESENT
2 -27.575771 152.989980 ... iNaturalist Australia PRESENT
3 -27.574852 152.992831 ... eBird Australia PRESENT
4 -27.574169 152.995529 ... eBird Australia PRESENT
... ... ... ... ... ...
2786 -27.517759 152.985278 ... eBird Australia PRESENT
2787 -27.517759 152.985278 ... eBird Australia PRESENT
2788 -27.517759 152.985278 ... eBird Australia PRESENT
2789 -27.517759 152.985278 ... eBird Australia PRESENT
2790 -27.517759 152.985278 ... eBird Australia PRESENT
[2791 rows x 8 columns]
>>> brisbane_parks_all.plot(edgecolor = "#5A5A5A", linewidth = 1, facecolor = "white", figsize = (7,10))
>>> plt.plot(*plot_brisbane_parks_bbox.exterior.xy,color="red")
>>> plt.ylabel("Latitude",size=16,x=.45,y=0.5)
>>> plt.xlabel("Longitude",size=16)
>>> plt.scatter(lorikeet_brisbane["decimalLongitude"],lorikeet_brisbane["decimalLatitude"],alpha=0.5,color="orange",label="Lorikeet occurrences")
>>> plt.legend(loc=(0.5,0.96))
>>> # Filter records down to only those in the shapefile polygons
>>> brisbane_parks["count"] = pd.Series([0 for i in range(len(brisbane_parks))])
>>> for i,park in brisbane_parks.iterrows():
... points = [(x,y) for x,y in zip(lorikeet_brisbane["decimalLongitude"], lorikeet_brisbane["decimalLatitude"]) if shapely.contains_xy(park["geometry"],x,y)]
... brisbane_parks.at[i,"count"] = len(points)
>>> brisbane_parks_counts = brisbane_parks[["PARK_NAME","count"]].sort_values("count",ascending=False).reset_index(drop=True)
>>> brisbane_parks_counts.head(10)
PARK_NAME count
0 NYUNDARE-BA PARK 552
1 SHERWOOD ARBORETUM 475
2 FAULKNER PARK 64
3 FORT ROAD BUSHLAND 46
4 STRICKLAND TERRACE PARK 44
5 BENARRAWA RESERVE 34
6 GRACEVILLE RIVERSIDE PARKLANDS 33
7 NOSWORTHY PARK 17
8 HORACE WINDOW RESERVE 14
9 GRACEVILLE AVENUE PARK 7
Some shapefiles cover large geographic areas with the caveat that even the bounding box doesn’t restrict the number of records to a value that can be downloaded easily. In this case, we recommend more nuances and detailed methods that can be performed using looping techniques. One of our ALA Labs blog posts, Hex maps for species occurrence data, has been written detailing how to approach larger problems such as this.
